GenSeg: GenAI for Image Segmentaton
Semantic segmentation of medical images is pivotal in applications like disease diagnosis and treatment planning. While deep learning automates this task effectively, it struggles in ultra low-data regimes for the scarcity of annotated segmentation masks. To address this, we propose a generative deep learning framework that produces high-quality image-mask pairs as auxiliary training data.
Unlike traditional generative models that separate data generation from model training, ours uses multi-level optimization for end-to-end data generation. This allows segmentation performance to guide the generation process, producing data tailored to improve segmentation outcomes. Our method demonstrates strong generalization across 11 medical image segmentation tasks and 19 datasets, covering various diseases, organs, and modalities. It improves performance by 10–20% (absolute) in both same- and out-of-domain settings and requires 8–20 times less training data than existing approaches. This greatly enhances the feasibility and cost-effectiveness of deep learning in data-limited medical imaging scenarios.
Introduction
Medical image semantic segmentation is a pivotal process in the modern healthcare landscape, playing an indispensable role in diagnosing diseases, tracking disease progression, planning treatments, assisting surgeries, and supporting numerous other clinical activities. This process involves classifying each pixel within a specific image, such as a skin dermoscopy image, with a corresponding semantic label, such as skin cancer or normal skin.
The advent of deep learning has revolutionized this domain, offering unparalleled precision and automation in the segmentation of medical images. Despite these advancements, training accurate and robust deep learning models requires extensive, annotated medical imaging datasets, which are notoriously difficult to obtain. Labeling semantic segmentation masks for medical images is both time-intensive and costly, as it necessitates annotating each pixel. It requires not only substantial human resources but also specialized domain expertise. This leads to what is termed as ultra low-data regimes—scenarios where the availability of annotated training images is remarkably scarce. This scarcity poses a substantial challenge to the existing deep learning methodologies, causing them to overfit to training data and exhibit poor generalization performance on test images.
To address the scarcity of labeled image-mask pairs in semantic segmentation, several strategies have been devised, including data augmentation and semi-supervised learning approaches. Data augmentation techniques create synthetic pairs of images and masks, which are then utilized as supplementary training data. A significant limitation of these methods is that they treat data augmentation and segmentation model training as separate activities. Consequently, the process of data augmentation is not influenced by segmentation performance, leading to a situation where the augmented data might not contribute effectively to enhancing the model’s segmentation capabilities. Semi-supervised learning techniques exploit additional, unlabeled images to bolster segmentation accuracy. Despite their potential, these methods face limitations due to the necessity for extensive volumes of unlabeled images, a requirement often difficult to fulfill in medical settings where even unlabeled data can be challenging to obtain due to privacy issues, regulatory hurdles (e.g., IRB approvals), among others. Recent advancements in generative deep learning have opened new possibilities for overcoming such challenges by generating synthetic data. Compared to traditional augmentation methods, generative models have the potential to produce more realistic and diverse samples. However, most existing data generation or augmentation approaches do not incorporate feedback from the segmentation performance itself. Some recent studies have proposed multi-level optimization (MLO) frameworks in which the data generation process is guided by downstream tasks, such as classification. Yet, applying such optimization effectively to segmentation tasks remains underexplored. Moreover, unlike semi-supervised segmentation methods, generative approaches have the advantage of not requiring additional unlabeled data—an important benefit in sensitive medical domains.
In this work, we introduce GenSeg, a generative deep learning framework designed to address the challenges of ultra low-data regimes in medical image segmentation. GenSeg generates high-fidelity paired segmentation masks and medical images through a MLO process directly guided by segmentation performance. This ensures that the generated data not only meets high-quality standards but is also optimized to improve downstream model training. Unlike existing augmentation methods, GenSeg performs end-to-end data generation tightly coupled with segmentation objectives; unlike semi-supervised approaches, it requires no additional unlabeled images. GenSeg is a versatile, model-agnostic framework that can be seamlessly integrated into existing segmentation pipelines. We validated GenSeg across 11 segmentation tasks and 19 datasets spanning diverse imaging modalities, diseases, and organs. When integrated with UNet and DeepLab, GenSeg significantly boosts performance in ultra low-data settings (e.g., using only 50 training examples), achieving absolute gains of 10–20% in both same-domain and out-of-domain (OOD) generalization. Additionally, GenSeg demonstrates strong data efficiency, matching or exceeding baseline performance while requiring 8–20× fewer labeled samples.
Results
GenSeg overview
GenSeg is an end-to-end data generation framework designed to generate high-quality, labeled data, to enable the training of accurate medical image segmentation models in ultra low-data regimes (Fig. 1a). Our framework integrates two components: a data generation model and a semantic segmentation model. The data generation model is responsible for generating synthetic pairs of medical images and their corresponding segmentation masks. This generated data serves as the training material for the segmentation model. In our data generation process, we introduce a reverse generation mechanism. This mechanism initially generates segmentation masks, and subsequently, medical images, adhering to a progression from simpler to more complex tasks. Specifically, given an expert-annotated real segmentation mask, we apply basic image augmentation operations to produce an augmented mask, which is then inputted into a deep generative model to generate the corresponding medical image. A key distinction of our method lies in the architecture of this generative model. Unlike traditional models that rely on manually designed architecture, our model automatically learns this architecture from data (Fig. 1b, c). This adaptive architecture enables more nuanced and effective generation of medical images, tailored to the specific characteristics of the augmented segmentation masks.
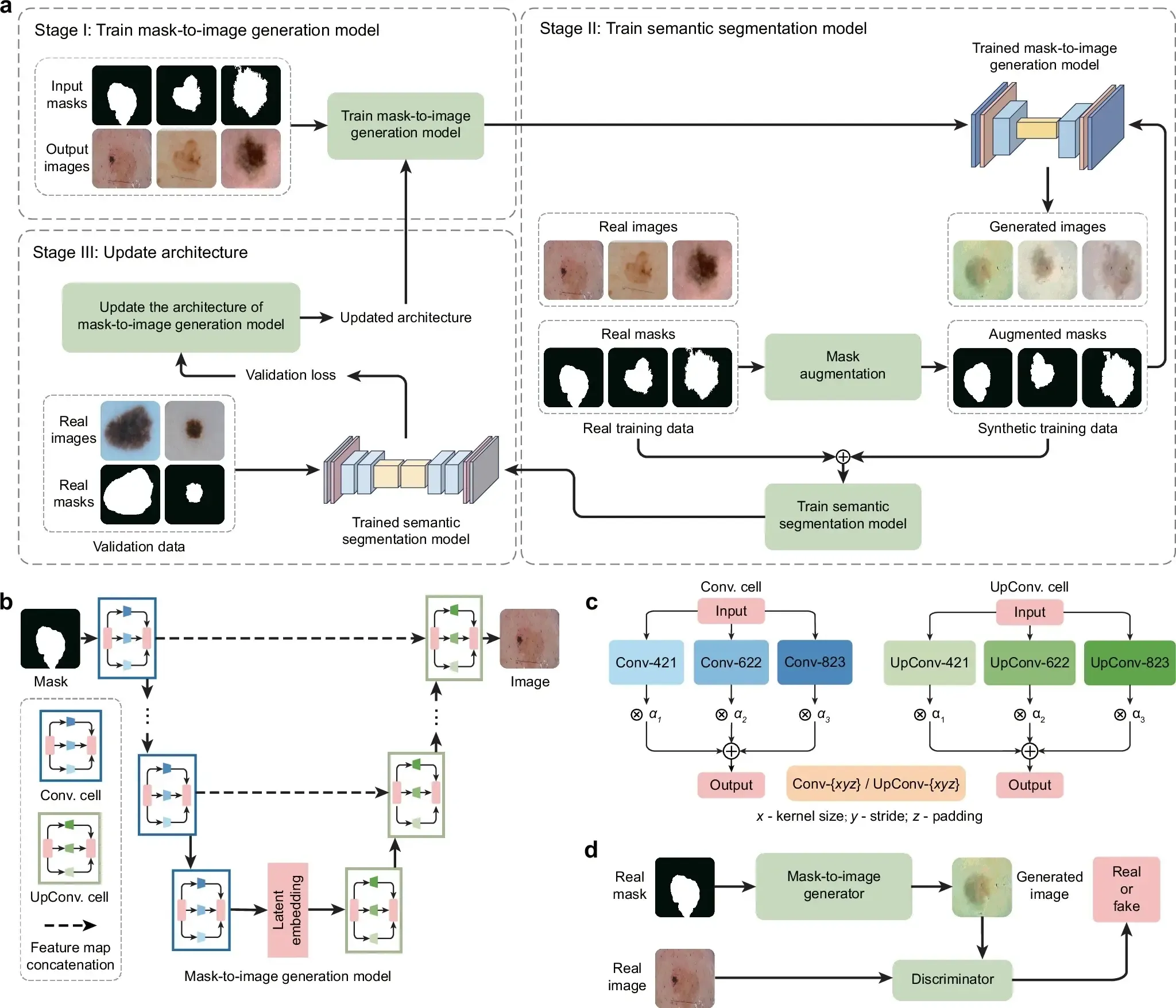
a Overview of the GenSeg framework. GenSeg consists of (1) a semantic segmentation model that predicts a segmentation mask from an input image, and (2) a mask-to-image generation model that synthesizes an image from a segmentation mask. The latter features a neural architecture that is both learnable in structure and parameterized by trainable weights. GenSeg operates through three end-to-end learning stages. In stage I, the network weights of the mask-to-image model are trained with real mask-image pairs, with its architecture tentatively fixed. Stage II involves using the trained mask-to-image model to synthesize training data. Real segmentation masks are augmented to create new masks, from which synthetic images are generated. These synthetic image-mask pairs are used alongside real data to train the segmentation model. In stage III, the trained segmentation model is evaluated on a real validation dataset, and the resulting validation loss—which reflects the performance of the mask-to-image model—is used to update this architecture. Following this update, the model re-enters Stage I for further training, and this cycle continues until convergence. b Searchable architecture of the mask-to-image generation model. It comprises an encoder and a decoder. The encoder processes an input mask into a latent representation using a series of searchable convolution (Conv.) cells. The decoder employs a stack of searchable up-convolution (UpConv.) cells to transform the latent representation into an output medical image. Each cell, as shown in (c) contains multiple candidate operations characterized by varying kernel sizes, strides, and padding options. Each operation is associated with a weight α denoting its importance. The architecture search process optimizes these weights, and only the most influential operations are retained in the final model. d The weight parameters of the mask-to-image generator are trained within a generative adversarial network (GAN) framework, in which a discriminator learns to distinguish real images from generated ones, while the generator is optimized to produce images that are indistinguishable from real images. All qualitative examples are sourced from publicly available datasets.
GenSeg features an end-to-end data generation strategy, which ensures a synergistic relationship between the generation of data and the performance of the segmentation model. By closely aligning the data generation process with the needs and feedback of the segmentation model, GenSeg ensures the relevance and utility of the generated data for effective training of the segmentation model. To evaluate the effectiveness of the generated data, we first train a semantic segmentation model using this data. We then assess the model’s performance on a validation set consisting of real medical images, each accompanied by an expert-annotated segmentation mask. The model’s validation performance serves as a reflection of the quality of the generated data: if the data is of low quality, the segmentation model trained on it will show poor performance during validation. By concentrating on improving the model’s validation performance, we can, in turn, enhance the quality of the generated data.
Our approach utilizes a MLO strategy to achieve end-to-end data generation. MLO involves a series of nested optimization problems, where the optimal parameters from one level serve as inputs for the objective function at the next level. Conversely, parameters that are not yet optimized at a higher level are fed back as inputs to lower levels. This yields a dynamic, iterative process that solves optimization problems in different levels jointly. Our method employs a three-tiered MLO process, executed end-to-end. The first level focuses on training the weight parameters of our data generation model, while keeping its learnable architecture constant. This training is performed within a generative adversarial network (GAN) framework (Fig. 1d), where a discriminator network learns to distinguish between real and generated images, and the data generation model is optimized to fool the discriminator by producing images that closely resemble real ones. At the second level, this trained model is used to produce synthetic image-mask pairs, which are then employed to train a semantic segmentation model. The final level involves validating the segmentation model using real medical images with expert-annotated masks. The performance of the segmentation model in this validation phase is a function of the architecture of the generation model. We optimize this architecture by minimizing the validation loss. By jointly solving the three levels of nested optimization problems, we can concurrently train data generation and semantic segmentation models in an end-to-end manner.
Our framework was validated for a variety of medical imaging segmentation tasks across 19 datasets, spanning a diverse spectrum of imaging techniques, diseases, lesions, and organs. These tasks comprise segmentation of skin lesions from dermoscopy images, breast cancer from ultrasound images, placental vessels from fetoscopic images, polyps from colonoscopy images, foot ulcers from standard camera images, intraretinal cystoid fluid from optical coherence tomography (OCT) images, lungs from chest X-ray images, and left ventricles and myocardial wall from echocardiography images.
GenSeg performance
We evaluated GenSeg’s performance in ultra-low data regimes. We conducted three independent runs for each dataset using different random seeds. The reported results represent the mean and standard deviation computed across these runs. GenSeg, being a versatile framework, facilitates training various backbone segmentation models with its generated data. To demonstrate this versatility, we applied GenSeg to two popular models: UNet and DeepLab, resulting in GenSeg-UNet and GenSeg-DeepLab, respectively. GenSeg-DeepLab and GenSeg-UNet demonstrated significant performance improvements over DeepLab and UNet in scenarios with limited data (Fig. 2a and Supplementary Fig. 1). Specifically, in the tasks of segmenting placental vessels, skin lesions, polyps, intraretinal cystoid fluids, foot ulcers, and breast cancer, with training sets as small as 50, 40, 40, 50, 50, and 100 samples respectively, GenSeg-DeepLab outperformed DeepLab substantially, with absolute percentage gains of 20.6%, 14.5%, 11.3%, 11.3%, 10.9%, and 10.4%. Similarly, GenSeg-UNet surpassed UNet by significant margins, recording absolute percentage improvements of 15%, 9.6%, 11%, 6.9%, 19%, and 12.6% across these tasks. The limited size of these training datasets presents significant challenges for accurately training DeepLab and UNet models. For example, DeepLab’s effectiveness in these tasks is limited, with performance varying from 0.31 to 0.62, averaging 0.51. In contrast, using our method, the performance significantly improves, ranging from 0.51 to 0.73 and averaging 0.64. This highlights the strong capability of our approach to achieve precise segmentation in ultra low-data regimes. Moreover, these segmentation tasks are highly diverse. For example, placental vessels involve complex branching structures, skin lesions vary in shape and size, and polyps require differentiation from surrounding mucosal tissue. GenSeg demonstrated robust performance enhancements across these diverse tasks, underscoring its strong capability in achieving accurate segmentation across different diseases, organs, and imaging modalities.
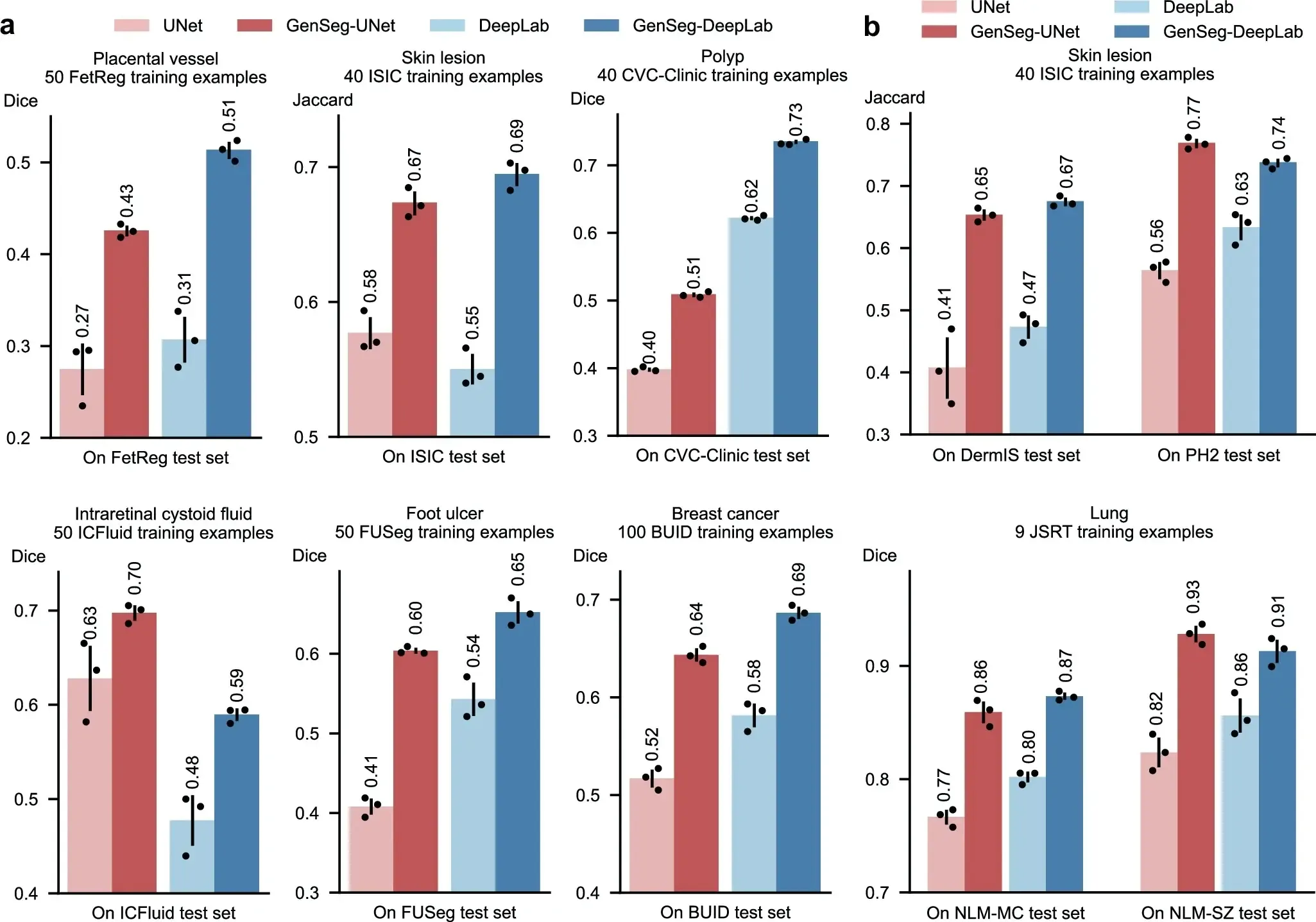
a The performance of GenSeg applied to UNet (GenSeg-UNet) and DeepLab (GenSeg-DeepLab) under in-domain settings (test and training data are from the same domain) in the tasks of segmenting placental vessels, skin lesions, polyps, intraretinal cystoid fluids, foot ulcers, and breast cancer using limited training data (50, 40, 40, 50, 50, and 100 examples from the FetReg, ISIC, CVC-Clinic, ICFluid, FUSeg, and BUID datasets, respectively for each task), compared to vanilla UNet and DeepLab. b The performance of GenSeg-UNet and GenSeg-DeepLab under out-of-domain settings (test and training data are from different domains) in segmenting skin lesions (using only 40 examples from the ISIC dataset for training, and the DermIS and PH2 datasets for testing) and lungs (using only 9 examples from the JSRT dataset for training, and the NLM-MC and NLM-SZ datasets for testing), compared to vanilla UNet and DeepLab. In all panels, bar heights represent the mean, and error bars indicate the standard deviation across three independent runs with different random seeds. Results from individual runs are shown as dot points. Source data are provided as a Source Data file.
Discussion
We present GenSeg, a robust data generation tool designed for generating high-quality data to enhance the training of medical image segmentation models. Demonstrating superior in-domain and OOD generalization performance across nine diverse segmentation tasks and 19 datasets, GenSeg excels particularly in scenarios with a limited number of real, expert-annotated training examples (as few as 50). GenSeg substantially enhances sample efficiency, requiring far fewer expert-annotated training examples than baseline methods to achieve similar performance. This greatly reduces both the burden and costs associated with medical image annotation.
GenSeg stands out by requiring fewer expert-annotated real training examples compared to baseline methods, yet it achieves comparable performance. This substantial reduction in the need for manually labeled segmentation masks significantly cuts down both the burden and costs associated with medical image annotation. With just a small set of real examples, GenSeg effectively trains a data generation model which then produces additional synthetic data, effectively mimicking the benefits of using a large dataset of real examples.
GenSeg significantly improves segmentation models’ OOD generalization capability. GenSeg is capable of generating diverse medical images accompanied by precise segmentation masks. When trained on this diverse augmented dataset, segmentation models can learn more robust and OOD generalizable feature representations.
GenSeg stands out from current data augmentation and generation techniques by offering superior segmentation performance, primarily due to its end-to-end data generation mechanism. Unlike previous methods that separate data augmentation/generation and segmentation model training, our approach integrates them end-to-end within a unified, MLO framework. Within this framework, the validation performance of the segmentation model acts as a direct indicator of the generated data’s usefulness. By leveraging this performance to inform the training process of the generation model, we ensure that the data produced is specifically optimized to improve the segmentation model. In previous methods, segmentation performance does not impact the process of data augmentation and generation. As a result, the augmented/generated data might not be effectively tailored for training the segmentation model. Furthermore, our framework learns a generative model that excels in generating data with greater diversity compared to existing augmentation methods.
GenSeg excels in surpassing semi-supervised segmentation methods without the need for external unlabeled images. In the context of medical imaging, collecting even unlabeled images presents a significant challenge due to stringent privacy concerns and regulatory constraints (e.g., IRB approval), thereby reducing the feasibility of semi-supervised methods. Despite the use of unlabeled real images, semi-supervised approaches underperform compared to GenSeg. This is primarily because these methods struggle to generate accurate masks for unlabeled images, meaning they are less effective at creating labeled training data. On the other hand, GenSeg is capable of producing high-quality images from masks, ensuring a close correspondence between the images’ content and the masks, thereby efficiently generating labeled training examples.
Our framework is designed to be universally applicable and independent of specific models. This design choice enables it to augment the capabilities of a broad spectrum of semantic segmentation models. To apply our framework to a specific segmentation model, the only requirement is to integrate the segmentation model into the second and third stages of our framework. This straightforward process enables researchers and practitioners to easily utilize our approach to improve the performance of diverse semantic segmentation models.
GenSeg presents several limitations that warrant attention. First, although GenSeg generates high-quality synthetic image-mask pairs, its performance may still be dependent on the quality and diversity of the limited real-world training data available. If the small dataset used to guide the generation process is highly biased or unrepresentative, the synthetic data produced may inherit these biases, potentially leading to suboptimal generalization on unseen cases. Additionally, while GenSeg demonstrates strong OOD performance, its generalization capabilities may diminish when faced with divergent datasets or imaging modalities that differ significantly from the training set. Furthermore, although GenSeg does not require extensive unlabeled data like semi-supervised methods, it still relies on a small set of expert-annotated data to initiate the synthetic data generation process, meaning that its utility may be limited in cases where even a small annotated dataset is difficult to obtain. Finally, the integration of GenSeg into clinical workflows would require validation in real-world settings to ensure that the synthetic data does not introduce artifacts or inconsistencies that could affect diagnostic decisions. Addressing these limitations in future iterations of GenSeg would be crucial for broadening its applicability and improving its robustness in diverse clinical environments.
Future research on GenSeg can progress in multiple directions. A key area is improving synthetic data generation to better represent complex anatomical structures and the variability inherent in diverse imaging modalities. This could involve refining the MLO process to capture finer details or incorporating advanced neural architectures to enhance the quality of synthetic images. Additionally, using generative models that can learn from limited examples may help GenSeg generalize more effectively across a broader range of medical scenarios. Another important direction is applying domain adaptation techniques to improve GenSeg’s robustness when encountering datasets that diverge significantly from the training data, such as novel imaging technologies or underrepresented patient populations. This would ensure more reliable performance in real-world clinical settings. Extending GenSeg’s capabilities beyond segmentation to tackle other medical imaging challenges, like anomaly detection, image registration, or multimodal image fusion, could further expand its utility. Such developments would position GenSeg as a more versatile tool for medical image analysis, addressing a wider array of diagnostic and treatment planning needs. Furthermore, integrating feedback from clinical experts into the synthetic data generation process could increase its clinical relevance, aligning outputs more closely with diagnostic practices. These research directions could enhance GenSeg’s adaptability and effectiveness across diverse medical imaging task.
An important consideration in evaluating the realism and utility of generated masks is how their variability compares to inter-reader variability observed in expert annotations. While our current study does not include a direct comparison—due to the use of datasets with only a single reference annotation per image—this is a valuable direction for future work. Qualitatively, we find that the augmented masks produced by our generative model exhibit anatomically plausible and semantically consistent variations, often resembling the natural diversity seen across patients and imaging conditions. Quantitatively, the consistent improvements in segmentation accuracy suggest that these synthetic masks enrich the training set with meaningful variability. Nevertheless, a systematic comparison with inter-reader variability would provide deeper insights into the clinical realism of the generated data. Incorporating multi-reader datasets in future evaluations could help assess whether the diversity introduced by generative augmentation aligns with the range of acceptable expert interpretations.
In summary, GenSeg is a robust data generation tool that seamlessly integrates with current semantic segmentation models. It significantly enhances both in-domain and OOD generalization performance in ultra low-data regimes, markedly boosting sample efficiency. Furthermore, it surpasses state-of-the-art methods in data augmentation and semi-supervised learning.